Note
Click here to download the full example code
Tutorial for multilabel-classification¶
In this tutorial, we compare the prediction sets estimated by the RCPS and CRC methods implemented in MAPIE, for recall control purpose, on a two-dimensional toy dataset. We will also look at the Learn Then Test (LTT) procedure. It allows to create prediction sets for precision control.
Throughout this tutorial, we will answer the following questions:
How does the threshold vary according to the desired risk?
Is the chosen conformal method well calibrated (i.e. does the actual risk equal to the desired one) ?
import matplotlib.pyplot as plt
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.multioutput import MultiOutputClassifier
from sklearn.naive_bayes import GaussianNB
from mapie.multi_label_classification import MapieMultiLabelClassifier
1. Construction of the dataset¶
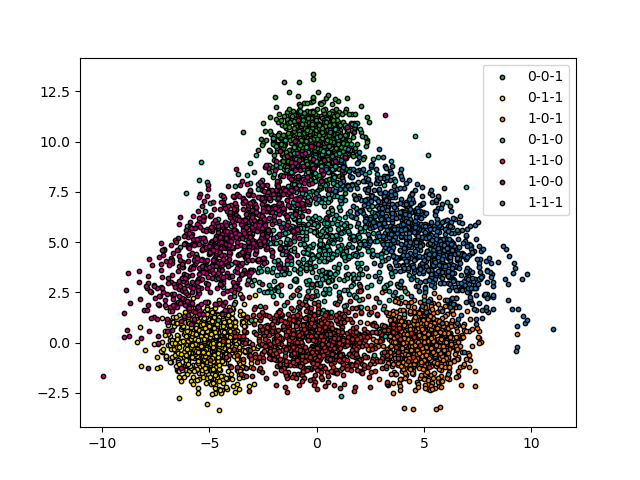
We use a two-dimensional toy dataset with three possible labels. The idea is to create a triangle where the observations on the edges have only one label, those on the vertices have two labels (those of the two edges) and the center have all the labels
centers = [(0, 10), (-5, 0), (5, 0), (0, 5), (0, 0), (-4, 5), (5, 5)]
covs = [
np.eye(2), np.eye(2), np.eye(2), np.diag([5, 5]), np.diag([3, 1]),
np.array([
[4, 3],
[3, 4]
]),
np.array([
[3, -2],
[-2, 3]
]),
]
x_min, x_max, y_min, y_max, step = -15, 15, -5, 15, 0.1
n_samples = 800
X = np.vstack([
np.random.multivariate_normal(center, cov, n_samples)
for center, cov in zip(centers, covs)
])
classes = [
[1, 0, 1], [1, 1, 0], [0, 1, 1], [1, 1, 1],
[0, 1, 0], [1, 0, 0], [0, 0, 1]
]
y = np.vstack([np.full((n_samples, 3), row) for row in classes])
X_train_cal, X_test, y_train_cal, y_test = train_test_split(
X, y, test_size=0.2
)
X_train, X_cal, y_train, y_cal = train_test_split(
X_train_cal, y_train_cal, test_size=0.25
)
Let’s see our data.
colors = {
(0, 0, 1): {"color": "#1f77b4", "lac": "0-0-1"},
(0, 1, 1): {"color": "#ff7f0e", "lac": "0-1-1"},
(1, 0, 1): {"color": "#2ca02c", "lac": "1-0-1"},
(0, 1, 0): {"color": "#d62728", "lac": "0-1-0"},
(1, 1, 0): {"color": "#ffd700", "lac": "1-1-0"},
(1, 0, 0): {"color": "#c20078", "lac": "1-0-0"},
(1, 1, 1): {"color": "#06C2AC", "lac": "1-1-1"}
}
for i in range(7):
plt.scatter(
X[n_samples * i:n_samples * (i + 1), 0],
X[n_samples * i:n_samples * (i + 1), 1],
color=colors[tuple(y[n_samples * i])]["color"],
marker='o',
s=10,
edgecolor='k'
)
plt.legend([c["lac"] for c in colors.values()])
plt.show()
2 Recall control risk with CRC and RCPS¶
2.1 Fitting MapieMultiLabelClassifier¶
MapieMultiLabelClassifier will be fitted with RCPS and CRC methods. For the RCPS method, we will test all three Upper Confidence Bounds (Hoeffding, Bernstein and Waudby-Smith–Ramdas). The two methods give two different guarantees on the risk:
RCPS:
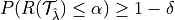
where 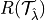 is the risk we want to control and
is the risk we want to control and  is the desired risk
is the desired risk
CRC:
![\mathbb{E}\left[L_{n+1}(\hat{\lambda})\right] \leq \alpha](../../_images/math/0978b747193b7d53782d308bcb680bb9c13522a1.png)
where 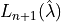 is the risk of a new observation and
is the risk of a new observation and
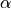 is the desired risk
is the desired risk
In both cases, the objective of the method is to find the optimal value of
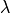 (threshold above which we consider a label as being present)
such that the recall on the test points is at least equal to the required
recall.
(threshold above which we consider a label as being present)
such that the recall on the test points is at least equal to the required
recall.
method_params = {
"RCPS - Hoeffding": ("rcps", "hoeffding"),
"RCPS - Bernstein": ("rcps", "bernstein"),
"RCPS - WSR": ("rcps", "wsr"),
"CRC": ("crc", None)
}
clf = MultiOutputClassifier(GaussianNB()).fit(X_train, y_train)
alpha = np.arange(0.01, 1, 0.01)
y_pss, recalls, thresholds, r_hats, r_hat_pluss = {}, {}, {}, {}, {}
y_test_repeat = np.repeat(y_test[:, :, np.newaxis], len(alpha), 2)
for i, (name, (method, bound)) in enumerate(method_params.items()):
mapie = MapieMultiLabelClassifier(
estimator=clf, method=method, metric_control="recall"
)
mapie.fit(X_cal, y_cal)
_, y_pss[name] = mapie.predict(
X_test, alpha=alpha, bound=bound, delta=.1
)
recalls[name] = (
(y_test_repeat * y_pss[name]).sum(axis=1) /
y_test_repeat.sum(axis=1)
).mean(axis=0)
thresholds[name] = mapie.lambdas_star
r_hats[name] = mapie.r_hat
r_hat_pluss[name] = mapie.r_hat_plus
2.2. Results¶
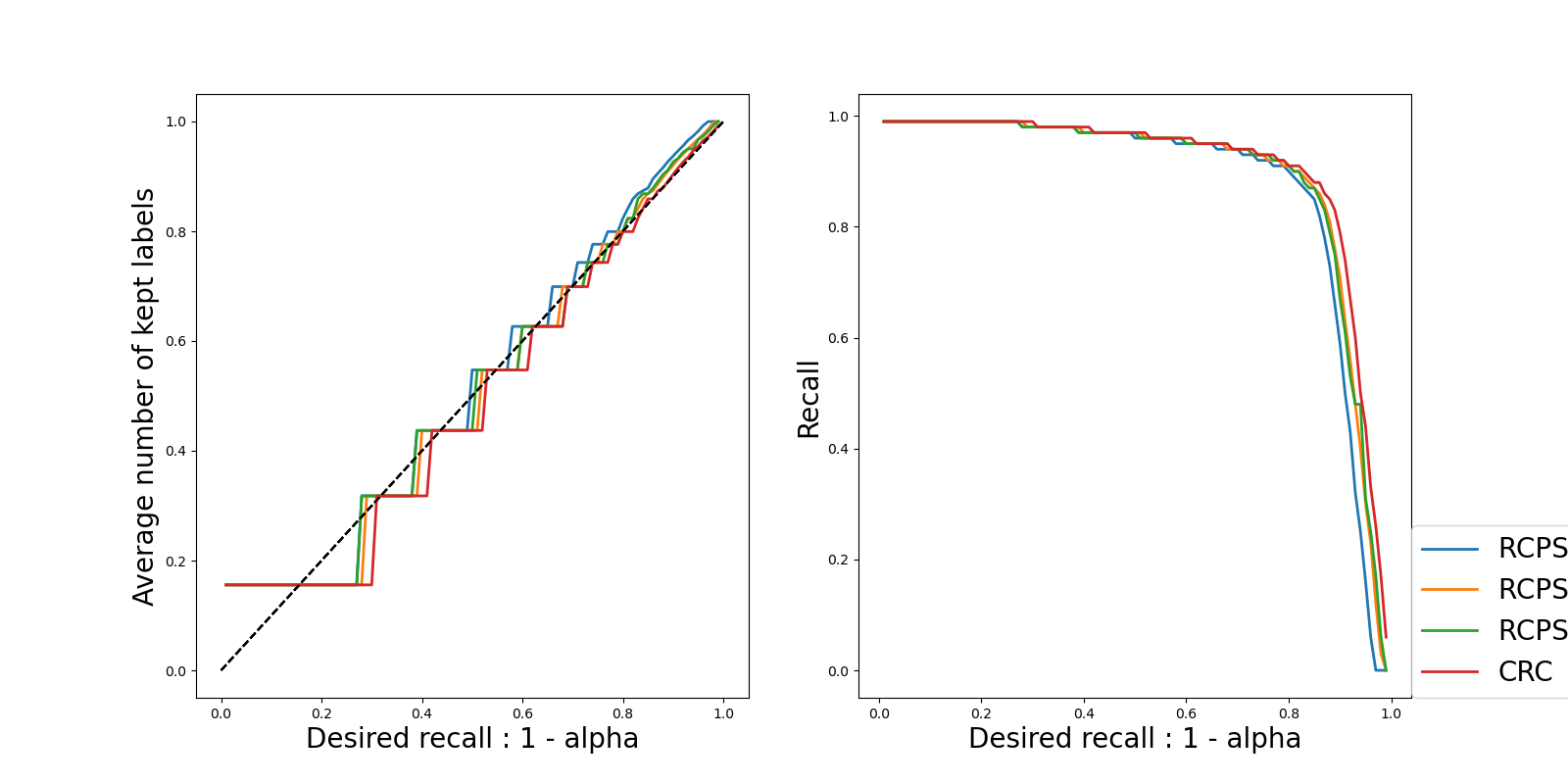
To check the results of the methods, we propose two types of plots:
Plots where the confidence level varies. Here two metrics are plotted for each method and for each UCB
The actual recall (which should be always near to the required one): we can see that they are close to each other.
The value of the threshold: we see that the threshold is decreasing as
 increases, which is what is expected because a
smaller threshold will give larger prediction sets, hence a larger
recall.
increases, which is what is expected because a
smaller threshold will give larger prediction sets, hence a larger
recall.
vars_y = [recalls, thresholds]
labels_y = ["Average number of kept labels", "Recall", "Threshold"]
fig, axs = plt.subplots(1, len(vars_y), figsize=(8*len(vars_y), 8))
for i, var in enumerate(vars_y):
for name, (method, bound) in method_params.items():
axs[i].plot(1 - alpha, var[name], label=name, linewidth=2)
if i == 0:
axs[i].plot([0, 1], [0, 1], ls="--", color="k")
axs[i].set_xlabel("Desired recall : 1 - alpha", fontsize=20)
axs[i].set_ylabel(labels_y[i], fontsize=20)
if i == (len(vars_y) - 1):
axs[i].legend(fontsize=20, loc=[1, 0])
plt.show()
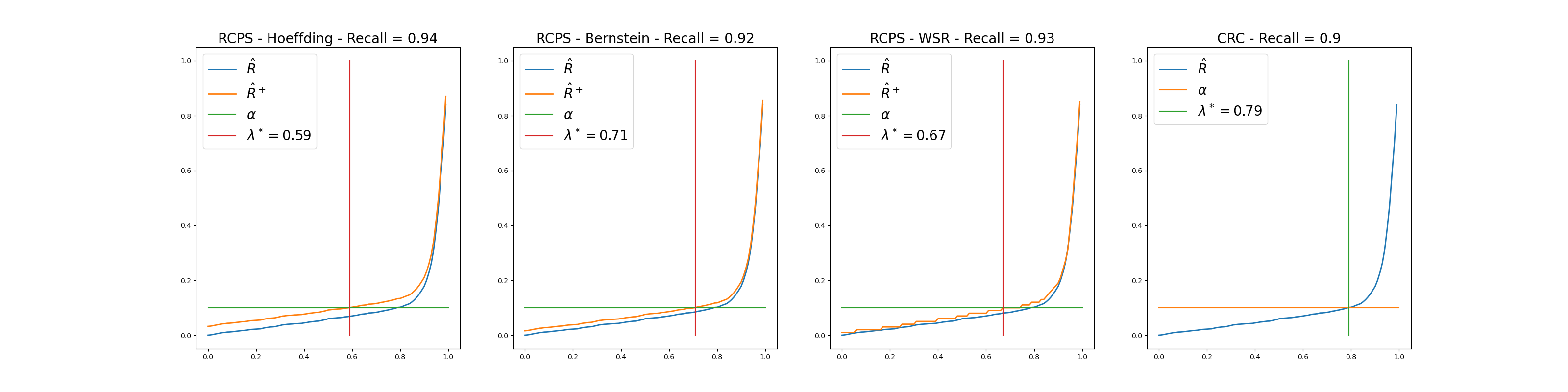
Plots where we choose a specific risk value (0.1 in our case) and look at the average risk, the UCB of the risk (for RCPS methods) and the choice of the threshold
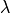
We can see that among the RCPS methods, the Bernstein method gives the best results as for a given value of
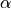 as we are above the required recall but with a larger value of
as we are above the required recall but with a larger value of
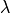 than the two others bounds.
than the two others bounds.The CRC method gives the best results since it guarantees the coverage with a larger threshold.
fig, axs = plt.subplots(
1,
len(method_params),
figsize=(8*len(method_params), 8)
)
for i, (name, (method, bound)) in enumerate(method_params.items()):
axs[i].plot(
mapie.lambdas,
r_hats[name], label=r"$\hat{R}$", linewidth=2
)
if name != "CRC":
axs[i].plot(
mapie.lambdas,
r_hat_pluss[name], label=r"$\hat{R}^+$", linewidth=2
)
axs[i].plot([0, 1], [alpha[9], alpha[9]], label=r"$\alpha$")
axs[i].plot(
[thresholds[name][9], thresholds[name][9]], [0, 1],
label=r"$\lambda^*" + f" = {thresholds[name][9]}$"
)
axs[i].legend(fontsize=20)
axs[i].set_title(
f"{name} - Recall = {round(recalls[name][9], 2)}",
fontsize=20
)
plt.show()
3. Precision control risk with LTT¶
3.1 Fitting MapieMultilabelClassifier¶
In this part, we will use LTT to control precision.
At the opposite of the 2 previous method, LTT can handle non-monotonous loss.
The procedure consist in multiple hypothesis testing. This is why the output
of this procedure isn’t reduce to one value of 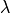 .
.
More precisely, we look after all the 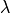 that sastisfy the
following:
that sastisfy the
following:
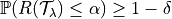 ,
where
,
where 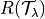 is the risk we want to control and
each
is the risk we want to control and
each 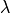 should satisfy FWER control.
should satisfy FWER control.
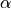 is the desired risk.
is the desired risk.
Notice that the procedure will diligently examine each 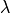 such that the risk remains below level
such that the risk remains below level 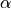 , meaning not
every
, meaning not
every 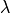 will be considered.
This means that a for a
will be considered.
This means that a for a 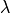 such that risk is below
such that risk is below
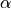 doesn’t necessarly pass the FWER control! This is what we are going to
explore.
doesn’t necessarly pass the FWER control! This is what we are going to
explore.
mapie_clf = MapieMultiLabelClassifier(
estimator=clf,
method='ltt',
metric_control='precision'
)
mapie_clf.fit(X_cal, y_cal)
alpha = 0.1
_, y_ps = mapie_clf.predict(
X_test,
alpha=alpha,
delta=0.1
)
valid_index = mapie_clf.valid_index[0] # valid_index is a list of list
lambdas = mapie_clf.lambdas[valid_index]
mini = lambdas[np.argmin(lambdas)]
maxi = lambdas[np.argmax(lambdas)]
r_hat = mapie_clf.r_hat
idx_max = np.argmin(r_hat[valid_index])
3.2 Valid parameters for precision control¶
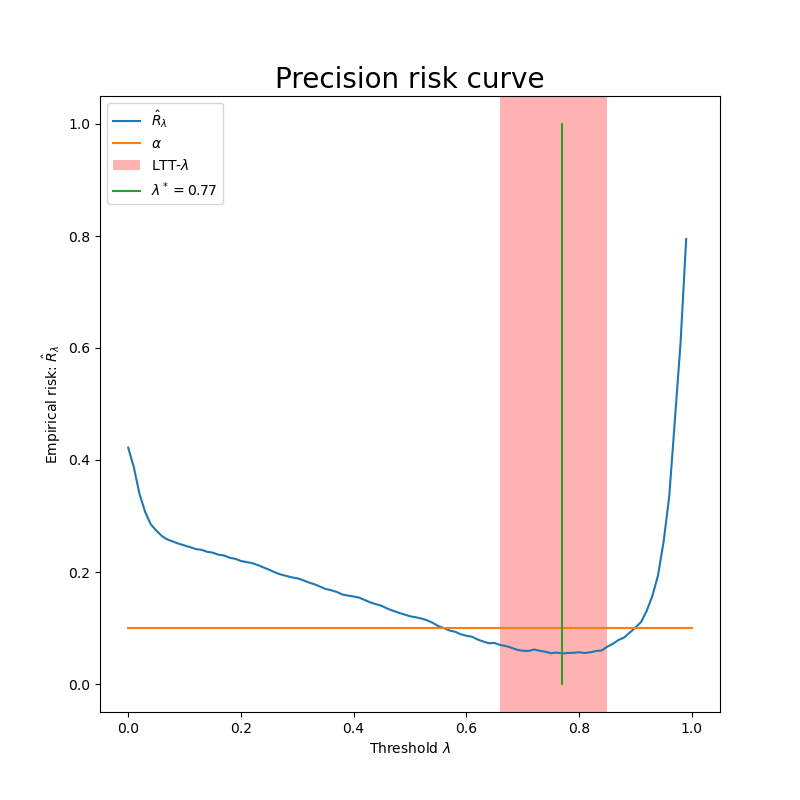
We can see that not all 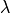 such that risk is below the orange
line are choosen by the procedure. Otherwise, all the lambdas that are
in the red rectangle verify family wise error rate control and allow to
control precision at the desired level with a high probability.
such that risk is below the orange
line are choosen by the procedure. Otherwise, all the lambdas that are
in the red rectangle verify family wise error rate control and allow to
control precision at the desired level with a high probability.
plt.figure(figsize=(8, 8))
plt.plot(mapie_clf.lambdas, r_hat, label=r"$\hat{R}_\lambda$")
plt.plot([0, 1], [alpha, alpha], label=r"$\alpha$")
plt.axvspan(mini, maxi, facecolor='red', alpha=0.3, label=r"LTT-$\lambda$")
plt.plot(
[lambdas[idx_max], lambdas[idx_max]], [0, 1],
label=r"$\lambda^* =" + f"{lambdas[idx_max]}$"
)
plt.xlabel(r"Threshold $\lambda$")
plt.ylabel(r"Empirical risk: $\hat{R}_\lambda$")
plt.title("Precision risk curve", fontsize=20)
plt.legend()
plt.show()
Total running time of the script: ( 0 minutes 2.265 seconds)