Note
Click here to download the full example code
Estimating prediction intervals of time series forecast with EnbPI¶
This example uses
MapieTimeSeriesRegressor to estimate
prediction intervals associated with time series forecast. It follows [6].
We use here the Victoria electricity demand dataset used in the book “Forecasting: Principles and Practice” by R. J. Hyndman and G. Athanasopoulos. The electricity demand features daily and weekly seasonalities and is impacted by the temperature, considered here as a exogeneous variable.
A Random Forest model is already fitted on data. The hyper-parameters are
optimized with a RandomizedSearchCV using a
sequential TimeSeriesSplit cross validation,
in which the training set is prior to the validation set.
The best model is then feeded into
MapieTimeSeriesRegressor to estimate the
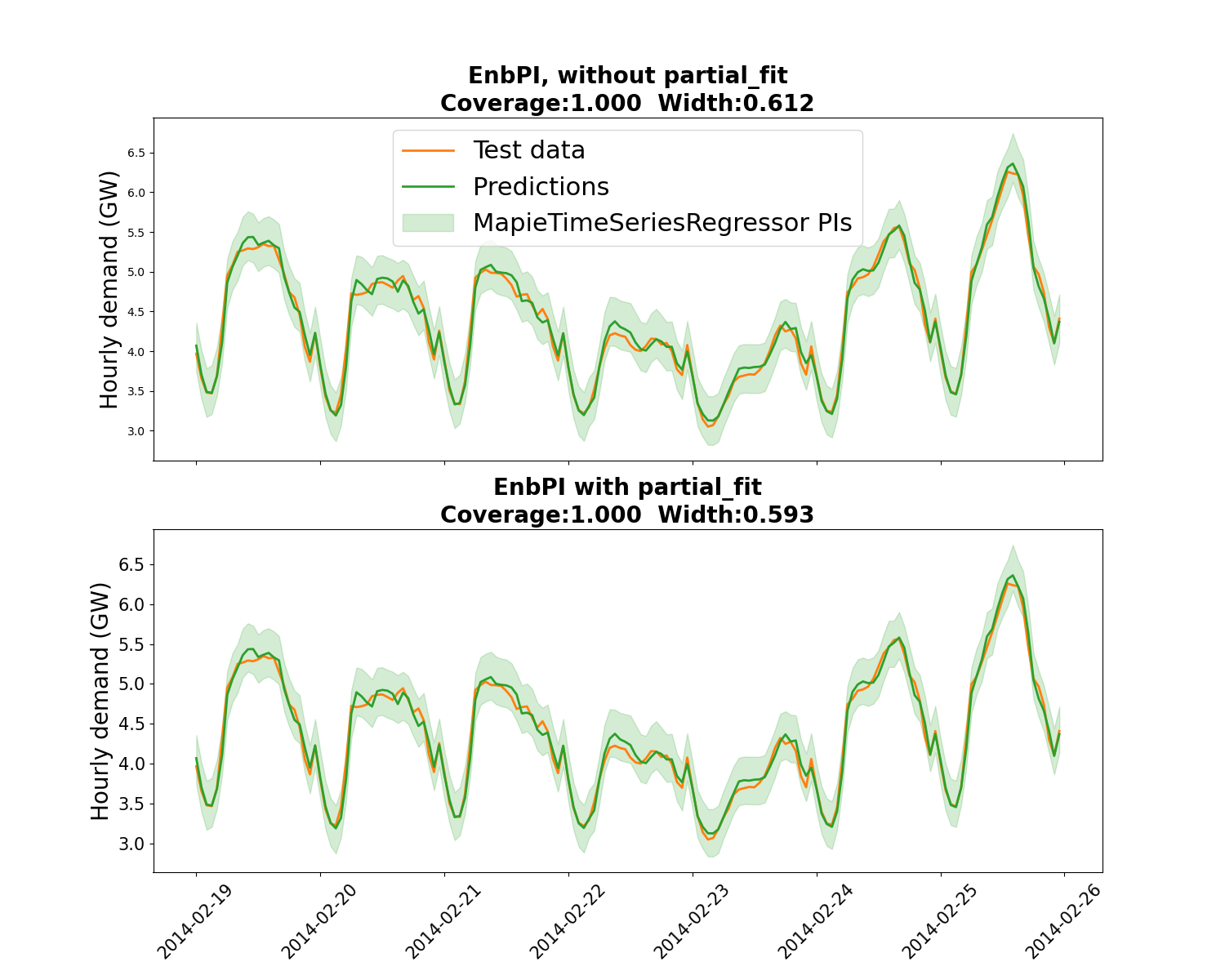
associated prediction intervals. We compare two approaches: with or without
partial_fit called at every step following [6]. It appears that
partial_fit offer a coverage closer to the targeted coverage, and with
narrower PIs.
Out:
EnbPI, with no partial_fit, width optimization
EnbPI with partial_fit, width optimization
Coverage / prediction interval width mean for MapieTimeSeriesRegressor:
EnbPI without any partial_fit:1.000, 0.612
Coverage / prediction interval width mean for MapieTimeSeriesRegressor:
EnbPI with partial_fit:1.000, 0.593
import warnings
from typing import cast
import numpy as np
import pandas as pd
from matplotlib import pylab as plt
from scipy.stats import randint
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import RandomizedSearchCV, TimeSeriesSplit
from mapie._typing import NDArray
from mapie.metrics import (regression_coverage_score,
regression_mean_width_score)
from mapie.regression import MapieTimeSeriesRegressor
from mapie.subsample import BlockBootstrap
warnings.simplefilter("ignore")
# Load input data and feature engineering
url_file = (
"https://raw.githubusercontent.com/scikit-learn-contrib/MAPIE/"
+ "master/examples/data/demand_temperature.csv"
)
demand_df = pd.read_csv(url_file, parse_dates=True, index_col=0)
demand_df["Date"] = pd.to_datetime(demand_df.index)
demand_df["Weekofyear"] = demand_df.Date.dt.isocalendar().week.astype("int64")
demand_df["Weekday"] = demand_df.Date.dt.isocalendar().day.astype("int64")
demand_df["Hour"] = demand_df.index.hour
n_lags = 5
for hour in range(1, n_lags):
demand_df[f"Lag_{hour}"] = demand_df["Demand"].shift(hour)
# Train/validation/test split
num_test_steps = 24 * 7
demand_train = demand_df.iloc[:-num_test_steps, :].copy()
demand_test = demand_df.iloc[-num_test_steps:, :].copy()
features = ["Weekofyear", "Weekday", "Hour", "Temperature"] + [
f"Lag_{hour}" for hour in range(1, n_lags)
]
X_train = demand_train.loc[
~np.any(demand_train[features].isnull(), axis=1), features
]
y_train = demand_train.loc[X_train.index, "Demand"]
X_test = demand_test.loc[:, features]
y_test = demand_test["Demand"]
perform_hyperparameters_search = False
if perform_hyperparameters_search:
# CV parameter search
n_iter = 100
n_splits = 5
tscv = TimeSeriesSplit(n_splits=n_splits)
random_state = 59
rf_model = RandomForestRegressor(random_state=random_state)
rf_params = {"max_depth": randint(2, 30), "n_estimators": randint(10, 100)}
cv_obj = RandomizedSearchCV(
rf_model,
param_distributions=rf_params,
n_iter=n_iter,
cv=tscv,
scoring="neg_root_mean_squared_error",
random_state=random_state,
verbose=0,
n_jobs=-1,
)
cv_obj.fit(X_train, y_train)
model = cv_obj.best_estimator_
else:
# Model: Random Forest previously optimized with a cross-validation
model = RandomForestRegressor(
max_depth=10, n_estimators=50, random_state=59
)
# Estimate prediction intervals on test set with best estimator
alpha = 0.05
cv_mapietimeseries = BlockBootstrap(
n_resamplings=10, n_blocks=10, overlapping=False, random_state=59
)
mapie_enpbi = MapieTimeSeriesRegressor(
model,
method="enbpi",
cv=cv_mapietimeseries,
agg_function="mean",
n_jobs=-1,
)
print("EnbPI, with no partial_fit, width optimization")
mapie_enpbi = mapie_enpbi.fit(X_train, y_train)
y_pred_npfit_enbpi, y_pis_npfit_enbpi = mapie_enpbi.predict(
X_test, alpha=alpha, ensemble=True, optimize_beta=True
)
coverage_npfit_enbpi = regression_coverage_score(
y_test, y_pis_npfit_enbpi[:, 0, 0], y_pis_npfit_enbpi[:, 1, 0]
)
width_npfit_enbpi = regression_mean_width_score(
y_pis_npfit_enbpi[:, 1, 0], y_pis_npfit_enbpi[:, 0, 0]
)
print("EnbPI with partial_fit, width optimization")
mapie_enpbi = mapie_enpbi.fit(X_train, y_train)
y_pred_pfit_enbpi = np.zeros(y_pred_npfit_enbpi.shape)
y_pis_pfit_enbpi = np.zeros(y_pis_npfit_enbpi.shape)
step_size = 1
(
y_pred_pfit_enbpi[:step_size],
y_pis_pfit_enbpi[:step_size, :, :],
) = mapie_enpbi.predict(
X_test.iloc[:step_size, :], alpha=alpha, ensemble=True, optimize_beta=True
)
for step in range(step_size, len(X_test), step_size):
mapie_enpbi.partial_fit(
X_test.iloc[(step - step_size):step, :],
y_test.iloc[(step - step_size):step],
)
(
y_pred_pfit_enbpi[step:step + step_size],
y_pis_pfit_enbpi[step:step + step_size, :, :],
) = mapie_enpbi.predict(
X_test.iloc[step:(step + step_size), :],
alpha=alpha,
ensemble=True,
optimize_beta=True,
)
coverage_pfit_enbpi = regression_coverage_score(
y_test, y_pis_pfit_enbpi[:, 0, 0], y_pis_pfit_enbpi[:, 1, 0]
)
width_pfit_enbpi = regression_mean_width_score(
y_pis_pfit_enbpi[:, 1, 0], y_pis_pfit_enbpi[:, 0, 0]
)
# Print results
print(
"Coverage / prediction interval width mean for MapieTimeSeriesRegressor: "
"\nEnbPI without any partial_fit:"
f"{coverage_npfit_enbpi :.3f}, {width_npfit_enbpi:.3f}"
)
print(
"Coverage / prediction interval width mean for MapieTimeSeriesRegressor: "
"\nEnbPI with partial_fit:"
f"{coverage_pfit_enbpi:.3f}, {width_pfit_enbpi:.3f}"
)
enbpi_no_pfit = {
"y_pred": y_pred_npfit_enbpi,
"y_pis": y_pis_npfit_enbpi,
"coverage": coverage_npfit_enbpi,
"width": width_npfit_enbpi,
}
enbpi_pfit = {
"y_pred": y_pred_pfit_enbpi,
"y_pis": y_pis_pfit_enbpi,
"coverage": coverage_pfit_enbpi,
"width": width_pfit_enbpi,
}
results = [enbpi_no_pfit, enbpi_pfit]
# Plot estimated prediction intervals on test set
fig, axs = plt.subplots(
nrows=2, ncols=1, figsize=(15, 12), sharex="col"
)
for i, (ax, w, result) in enumerate(
zip(axs, ["EnbPI, without partial_fit", "EnbPI with partial_fit"], results)
):
ax.set_ylabel("Hourly demand (GW)", fontsize=20)
ax.plot(demand_test.Demand, lw=2, label="Test data", c="C1")
ax.plot(
demand_test.index,
result["y_pred"],
lw=2,
c="C2",
label="Predictions",
)
y_pis = cast(NDArray, result["y_pis"])
ax.fill_between(
demand_test.index,
y_pis[:, 0, 0],
y_pis[:, 1, 0],
color="C2",
alpha=0.2,
label="MapieTimeSeriesRegressor PIs",
)
ax.set_title(
w + "\n"
f"Coverage:{result['coverage']:.3f} Width:{result['width']:.3f}",
fontweight="bold",
size=20
)
plt.xticks(size=15, rotation=45)
plt.yticks(size=15)
axs[0].legend(prop={'size': 22})
plt.show()
Total running time of the script: ( 0 minutes 39.352 seconds)