mapie.conformity_scores.AbsoluteConformityScore¶
- class mapie.conformity_scores.AbsoluteConformityScore(sym: bool = True)[source]¶
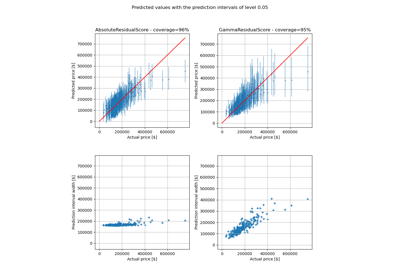
Absolute conformity score.
The signed conformity score = y - y_pred. The conformity score is symmetrical.
This is appropriate when the confidence interval is symmetrical and its range is approximatively the same over the range of predicted values.
- get_estimation_distribution(X: Union[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]], numpy._typing._nested_sequence._NestedSequence[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy._typing._nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]], y_pred: Union[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]], numpy._typing._nested_sequence._NestedSequence[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy._typing._nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]], conformity_scores: Union[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]], numpy._typing._nested_sequence._NestedSequence[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy._typing._nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]]) numpy.ndarray[Any, numpy.dtype[numpy._typing._array_like._ScalarType_co]][source]¶
Compute samples of the estimation distribution from the predicted values and the conformity scores, from the following formula: signed conformity score = y - y_pred <=> y = y_pred + signed conformity score
conformity_scorescan be either the conformity scores or the quantile of the conformity scores.
- get_signed_conformity_scores(X: Union[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]], numpy._typing._nested_sequence._NestedSequence[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy._typing._nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]], y: Union[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]], numpy._typing._nested_sequence._NestedSequence[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy._typing._nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]], y_pred: Union[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]], numpy._typing._nested_sequence._NestedSequence[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy._typing._nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]]) numpy.ndarray[Any, numpy.dtype[numpy._typing._array_like._ScalarType_co]][source]¶
Compute the signed conformity scores from the predicted values and the observed ones, from the following formula: signed conformity score = y - y_pred