mapie.regression.MapieTimeSeriesRegressor¶
- class mapie.regression.MapieTimeSeriesRegressor(estimator: Optional[sklearn.base.RegressorMixin] = None, method: str = 'enbpi', cv: Optional[Union[int, str, sklearn.model_selection._split.BaseCrossValidator]] = None, n_jobs: Optional[int] = None, agg_function: Optional[str] = 'mean', verbose: int = 0, conformity_score: Optional[mapie.conformity_scores.conformity_scores.ConformityScore] = None, random_state: Optional[Union[int, numpy.random.mtrand.RandomState]] = None)[source]¶
Prediction intervals with out-of-fold residuals for time series. This class only has two valid
method:"enbpi"or"aci"The prediction intervals are calibrated on a split of the trained data. Both strategies are estimating prediction intervals on single-output time series.
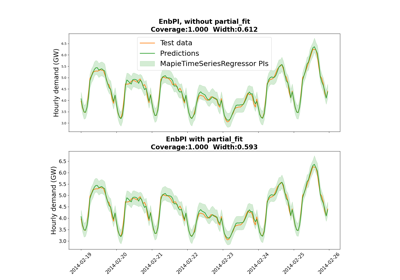
EnbPI allows you to update conformal scores using the
partial_fitfunction. It will replace the oldest one with the newest scores. It will keep the same amount of total scoresActually, EnbPI only corresponds to
MapieTimeSeriesRegressorif thecvargument is of typeBlockBootstrap.The ACI strategy allows you to adapt the conformal inference (i.e the quantile). If the real values are not in the coverage, the size of the intervals will grow. Conversely, if the real values are in the coverage, the size of the intervals will decrease. You can use a gamma coefficient to adjust the strength of the correction. If the quantile is equal to zero, the method will produce an infinite set size.
References
Chen Xu, and Yao Xie. “Conformal prediction for dynamic time-series.” https://arxiv.org/abs/2010.09107
Isaac Gibbs, Emmanuel Candes “Adaptive conformal inference under distribution shift” https://proceedings.neurips.cc/paper/2021/file/0d441de75945e5acbc865406fc9a2559-Paper.pdf
Margaux Zaffran et al. “Adaptive Conformal Predictions for Time Series” https://arxiv.org/pdf/2202.07282.pdf
- __init__(estimator: Optional[sklearn.base.RegressorMixin] = None, method: str = 'enbpi', cv: Optional[Union[int, str, sklearn.model_selection._split.BaseCrossValidator]] = None, n_jobs: Optional[int] = None, agg_function: Optional[str] = 'mean', verbose: int = 0, conformity_score: Optional[mapie.conformity_scores.conformity_scores.ConformityScore] = None, random_state: Optional[Union[int, numpy.random.mtrand.RandomState]] = None) None[source]¶
- adapt_conformal_inference(X: Union[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]], numpy._typing._nested_sequence._NestedSequence[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy._typing._nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]], y: Union[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]], numpy._typing._nested_sequence._NestedSequence[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy._typing._nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]], gamma: float, alpha: Optional[Union[float, Iterable[float]]] = None, ensemble: bool = False, optimize_beta: bool = False) mapie.regression.time_series_regression.MapieTimeSeriesRegressor[source]¶
Adapt the
alpha_tattribute when new data with known labels are available.- Parameters
- X: ArrayLike of shape (n_samples, n_features)
Input data.
- y: ArrayLike of shape (n_samples_test,)
Input labels.
- ensemble: bool
Boolean determining whether the predictions are ensembled or not. If
False, predictions are those of the model trained on the whole training set. IfTrue, predictions from perturbed models are aggregated by the aggregation function specified in theagg_functionattribute. Ifcvis"prefit"or"split",ensembleis ignored.By default
False.- gamma: float
Coefficient that decides the correction of the conformal inference. If it equals 0, there are no corrections.
- alpha: Optional[Union[float, Iterable[float]]]
Between
0and1, represents the uncertainty of the confidence interval.By default
None.- optimize_beta: bool
Whether to optimize the PIs’ width or not.
By default
False.
- Returns
- MapieTimeSeriesRegressor
The model itself.
- Raises
- ValueError
If the length of
yis greater than the length of the training set.
- partial_fit(X: Union[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]], numpy._typing._nested_sequence._NestedSequence[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy._typing._nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]], y: Union[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]], numpy._typing._nested_sequence._NestedSequence[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy._typing._nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]], ensemble: bool = False) mapie.regression.time_series_regression.MapieTimeSeriesRegressor[source]¶
Update the
conformity_scores_attribute when new data with known labels are available. Note: Don’t usepartial_fitwith samples of the training set.- Parameters
- X: ArrayLike of shape (n_samples_test, n_features)
Input data.
- y: ArrayLike of shape (n_samples_test,)
Input labels.
- ensemble: bool
Boolean determining whether the predictions are ensembled or not. If
False, predictions are those of the model trained on the whole training set. IfTrue, predictions from perturbed models are aggregated by the aggregation function specified in theagg_functionattribute. Ifcvis"prefit"or"split",ensembleis ignored.By default
False.
- Returns
- MapieTimeSeriesRegressor
The model itself.
- Raises
- ValueError
If the length of
yis greater than the length of the training set.
- predict(X: Union[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]], numpy._typing._nested_sequence._NestedSequence[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy._typing._nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]], ensemble: bool = False, alpha: Optional[Union[float, Iterable[float]]] = None, optimize_beta: bool = False, allow_infinite_bounds: bool = False) Union[numpy.ndarray[Any, numpy.dtype[numpy._typing._array_like._ScalarType_co]], Tuple[numpy.ndarray[Any, numpy.dtype[numpy._typing._array_like._ScalarType_co]], numpy.ndarray[Any, numpy.dtype[numpy._typing._array_like._ScalarType_co]]]][source]¶
Predict target on new samples with confidence intervals.
- Parameters
- X: ArrayLike of shape (n_samples, n_features)
Test data.
- ensemble: bool
Boolean determining whether the predictions are ensembled or not. If
False, predictions are those of the model trained on the whole training set. IfTrue, predictions from perturbed models are aggregated by the aggregation function specified in theagg_functionattribute. Ifcvis"prefit"or"split",ensembleis ignored.By default
False.- alpha: Optional[Union[float, Iterable[float]]]
Between
0and1, represents the uncertainty of the confidence interval.By default
None.- optimize_beta: bool
Whether to optimize the PIs’ width or not.
By default
False.- allow_infinite_bounds: bool
Allow infinite prediction intervals to be produced.
- Returns
- Union[NDArray, Tuple[NDArray, NDArray]]
NDArray of shape (n_samples,) if
alphaisNone.Tuple[NDArray, NDArray] of shapes (n_samples,) and (n_samples, 2, n_alpha) if
alphais notNone.[:, 0, :]: Lower bound of the prediction interval.
[:, 1, :]: Upper bound of the prediction interval.
- set_fit_request(*, groups: Union[bool, None, str] = '$UNCHANGED$', sample_weight: Union[bool, None, str] = '$UNCHANGED$') mapie.regression.time_series_regression.MapieTimeSeriesRegressor¶
Request metadata passed to the
fitmethod.Note that this method is only relevant if
enable_metadata_routing=True(seesklearn.set_config()). Please see User Guide on how the routing mechanism works.The options for each parameter are:
True: metadata is requested, and passed tofitif provided. The request is ignored if metadata is not provided.False: metadata is not requested and the meta-estimator will not pass it tofit.None: metadata is not requested, and the meta-estimator will raise an error if the user provides it.str: metadata should be passed to the meta-estimator with this given alias instead of the original name.
The default (
sklearn.utils.metadata_routing.UNCHANGED) retains the existing request. This allows you to change the request for some parameters and not others.New in version 1.3.
Note
This method is only relevant if this estimator is used as a sub-estimator of a meta-estimator, e.g. used inside a
pipeline.Pipeline. Otherwise it has no effect.- Parameters
- groupsstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
groupsparameter infit.- sample_weightstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
sample_weightparameter infit.
- Returns
- selfobject
The updated object.
- set_partial_fit_request(*, ensemble: Union[bool, None, str] = '$UNCHANGED$') mapie.regression.time_series_regression.MapieTimeSeriesRegressor¶
Request metadata passed to the
partial_fitmethod.Note that this method is only relevant if
enable_metadata_routing=True(seesklearn.set_config()). Please see User Guide on how the routing mechanism works.The options for each parameter are:
True: metadata is requested, and passed topartial_fitif provided. The request is ignored if metadata is not provided.False: metadata is not requested and the meta-estimator will not pass it topartial_fit.None: metadata is not requested, and the meta-estimator will raise an error if the user provides it.str: metadata should be passed to the meta-estimator with this given alias instead of the original name.
The default (
sklearn.utils.metadata_routing.UNCHANGED) retains the existing request. This allows you to change the request for some parameters and not others.New in version 1.3.
Note
This method is only relevant if this estimator is used as a sub-estimator of a meta-estimator, e.g. used inside a
pipeline.Pipeline. Otherwise it has no effect.- Parameters
- ensemblestr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
ensembleparameter inpartial_fit.
- Returns
- selfobject
The updated object.
- set_predict_request(*, allow_infinite_bounds: Union[bool, None, str] = '$UNCHANGED$', alpha: Union[bool, None, str] = '$UNCHANGED$', ensemble: Union[bool, None, str] = '$UNCHANGED$', optimize_beta: Union[bool, None, str] = '$UNCHANGED$') mapie.regression.time_series_regression.MapieTimeSeriesRegressor¶
Request metadata passed to the
predictmethod.Note that this method is only relevant if
enable_metadata_routing=True(seesklearn.set_config()). Please see User Guide on how the routing mechanism works.The options for each parameter are:
True: metadata is requested, and passed topredictif provided. The request is ignored if metadata is not provided.False: metadata is not requested and the meta-estimator will not pass it topredict.None: metadata is not requested, and the meta-estimator will raise an error if the user provides it.str: metadata should be passed to the meta-estimator with this given alias instead of the original name.
The default (
sklearn.utils.metadata_routing.UNCHANGED) retains the existing request. This allows you to change the request for some parameters and not others.New in version 1.3.
Note
This method is only relevant if this estimator is used as a sub-estimator of a meta-estimator, e.g. used inside a
pipeline.Pipeline. Otherwise it has no effect.- Parameters
- allow_infinite_boundsstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
allow_infinite_boundsparameter inpredict.- alphastr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
alphaparameter inpredict.- ensemblestr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
ensembleparameter inpredict.- optimize_betastr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
optimize_betaparameter inpredict.
- Returns
- selfobject
The updated object.
- set_score_request(*, sample_weight: Union[bool, None, str] = '$UNCHANGED$') mapie.regression.time_series_regression.MapieTimeSeriesRegressor¶
Request metadata passed to the
scoremethod.Note that this method is only relevant if
enable_metadata_routing=True(seesklearn.set_config()). Please see User Guide on how the routing mechanism works.The options for each parameter are:
True: metadata is requested, and passed toscoreif provided. The request is ignored if metadata is not provided.False: metadata is not requested and the meta-estimator will not pass it toscore.None: metadata is not requested, and the meta-estimator will raise an error if the user provides it.str: metadata should be passed to the meta-estimator with this given alias instead of the original name.
The default (
sklearn.utils.metadata_routing.UNCHANGED) retains the existing request. This allows you to change the request for some parameters and not others.New in version 1.3.
Note
This method is only relevant if this estimator is used as a sub-estimator of a meta-estimator, e.g. used inside a
pipeline.Pipeline. Otherwise it has no effect.- Parameters
- sample_weightstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
sample_weightparameter inscore.
- Returns
- selfobject
The updated object.
- update(X: Union[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]], numpy._typing._nested_sequence._NestedSequence[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy._typing._nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]], y: Union[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]], numpy._typing._nested_sequence._NestedSequence[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy._typing._nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]], ensemble: bool = False, alpha: Optional[Union[float, Iterable[float]]] = None, gamma: float = 0.0, optimize_beta: bool = False) mapie.regression.time_series_regression.MapieTimeSeriesRegressor[source]¶
Update with respect to the used
method.method="enbpi"will callpartial_fitmethod andmethod="aci"will calladapt_conformal_inferencemethod.- Parameters
- X: ArrayLike of shape (n_samples, n_features)
Input data.
- y: ArrayLike of shape (n_samples_test,)
Input labels.
- ensemble: bool
Boolean determining whether the predictions are ensembled or not. If
False, predictions are those of the model trained on the whole training set. IfTrue, predictions from perturbed models are aggregated by the aggregation function specified in theagg_functionattribute. Ifcvis"prefit"or"split",ensembleis ignored.By default
False.- alpha: Optional[Union[float, Iterable[float]]]
Between
0and1, represents the uncertainty of the confidence interval.By default
None.- gamma: float
Coefficient that decides the correction of the conformal inference. If it equals 0, there are no corrections.
By default
0..- optimize_beta: bool
Whether to optimize the PIs’ width or not.
By default
False.
- Returns
- MapieTimeSeriesRegressor
The model itself.
- Raises
- ValueError
If the length of
yis greater than the length of the training set.