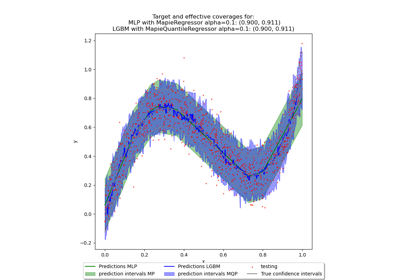
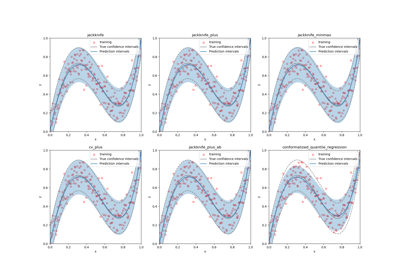
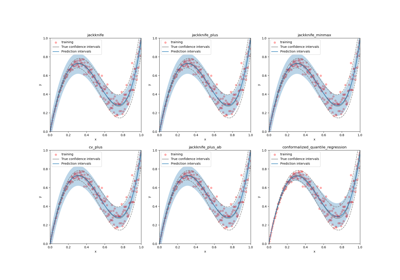
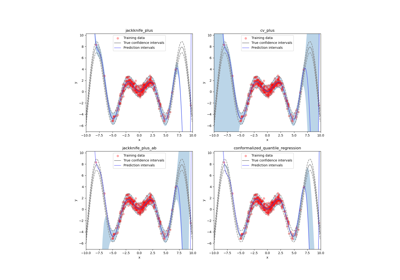
mapie.regression.MapieQuantileRegressor¶
- class mapie.regression.MapieQuantileRegressor(estimator: Optional[Union[sklearn.base.RegressorMixin, sklearn.pipeline.Pipeline, List[Union[sklearn.base.RegressorMixin, sklearn.pipeline.Pipeline]]]] = None, method: str = 'quantile', cv: Optional[str] = None, alpha: float = 0.1)[source]¶
This class implements the conformalized quantile regression strategy as proposed by Romano et al. (2019) to make conformal predictions. The only valid
methodis"quantile"and the only validcvis"split".- Parameters
- estimatorOptional[RegressorMixin]
Any regressor with scikit-learn API (i.e. with
fitandpredictmethods). IfNone, estimator defaults to aQuantileRegressorinstance.By default
"None".- method: str
Method to choose for prediction, in this case, the only valid method is the
"quantile"method.By default
"quantile".- cv: Optional[str]
The cross-validation strategy for computing conformity scores. In theory a split method is implemented as it is needed to provide both a training and calibration set.
By default
None.- alpha: float
Between
0.0and1.0, represents the risk level of the confidence interval. Loweralphaproduce larger (more conservative) prediction intervals.alphais the complement of the target coverage level.By default
0.1.
References
Yaniv Romano, Evan Patterson and Emmanuel J. Candès. “Conformalized Quantile Regression” Advances in neural information processing systems 32 (2019).
Examples
>>> import numpy as np >>> from mapie.regression import MapieQuantileRegressor >>> X_train = np.array([[0], [1], [2], [3], [4], [5]]) >>> y_train = np.array([5, 7.5, 9.5, 10.5, 12.5, 15]) >>> X_calib = np.array([[0], [1], [2], [3], [4], [5], [6], [7], [8], [9]]) >>> y_calib = np.array([5, 7, 9, 4, 8, 1, 5, 7.5, 9.5, 12]) >>> mapie_reg = MapieQuantileRegressor().fit( ... X_train, ... y_train, ... X_calib=X_calib, ... y_calib=y_calib ... ) >>> y_pred, y_pis = mapie_reg.predict(X_train) >>> print(y_pis[:, :, 0]) [[-8.16666667 19. ] [-6.33333333 20.83333333] [-4.5 22.66666667] [-2.66666667 24.5 ] [-0.83333333 26.33333333] [ 1. 28.16666667]] >>> print(y_pred) [ 5. 7. 9. 11. 13. 15.]
- Attributes
- valid_methods_: List[str]
List of all valid methods.
- single_estimator_: RegressorMixin
Estimator fitted on the whole training set.
- estimators_: List[RegressorMixin]
[0]: Estimator with quantile value of alpha/2
[1]: Estimator with quantile value of 1 - alpha/2
[2]: Estimator with quantile value of 0.5
- conformity_scores_: NDArray of shape (n_samples_train, 3)
Conformity scores between
y_calibandy_pred.[:, 0]: for
y_calibcoming from prediction estimator with quantile of alpha/2[:, 1]: for
y_calibcoming from prediction estimator with quantile of 1 - alpha/2[:, 2]: maximum of those first two scores
- n_calib_samples: int
Number of samples in the calibration dataset.
- __init__(estimator: Optional[Union[sklearn.base.RegressorMixin, sklearn.pipeline.Pipeline, List[Union[sklearn.base.RegressorMixin, sklearn.pipeline.Pipeline]]]] = None, method: str = 'quantile', cv: Optional[str] = None, alpha: float = 0.1) None[source]¶
- fit(X: Union[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]], numpy._typing._nested_sequence._NestedSequence[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy._typing._nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]], y: Union[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]], numpy._typing._nested_sequence._NestedSequence[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy._typing._nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]], sample_weight: Optional[Union[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]], numpy._typing._nested_sequence._NestedSequence[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy._typing._nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]]] = None, groups: Optional[Union[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]], numpy._typing._nested_sequence._NestedSequence[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy._typing._nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]]] = None, X_calib: Optional[Union[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]], numpy._typing._nested_sequence._NestedSequence[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy._typing._nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]]] = None, y_calib: Optional[Union[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]], numpy._typing._nested_sequence._NestedSequence[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy._typing._nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]]] = None, calib_size: Optional[float] = 0.3, random_state: Optional[Union[int, numpy.random.mtrand.RandomState]] = None, shuffle: Optional[bool] = True, stratify: Optional[Union[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]], numpy._typing._nested_sequence._NestedSequence[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy._typing._nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]]] = None, **fit_params) mapie.regression.quantile_regression.MapieQuantileRegressor[source]¶
Fit estimator and compute residuals used for prediction intervals. All the clones of the estimators for different quantile values are stored in order alpha/2, 1 - alpha/2, 0.5 in the
estimators_attribute. Residuals for the first two estimators and the maximum of residuals among these residuals are stored in theconformity_scores_attribute.- Parameters
- X: ArrayLike of shape (n_samples, n_features)
Training data.
- y: ArrayLike of shape (n_samples,)
Training labels.
- sample_weight: Optional[ArrayLike] of shape (n_samples,)
Sample weights for fitting the out-of-fold models. If
None, then samples are equally weighted. If some weights are null, their corresponding observations are removed before the fitting process and hence have no residuals. If weights are non-uniform, residuals are still uniformly weighted. Note that the sample weight defined are only for the training, not for the calibration procedure.By default
None.- groups: Optional[ArrayLike] of shape (n_samples,)
Always ignored, exists for compatibility.
- X_calib: Optional[ArrayLike] of shape (n_calib_samples, n_features)
Calibration data.
- y_calib: Optional[ArrayLike] of shape (n_calib_samples,)
Calibration labels.
- calib_size: Optional[float]
If
X_calibandy_calibare not defined, then the calibration dataset is created with the split defined bycalib_size.- random_state: Optional[Union[int, np.random.RandomState]], default=None
For the
sklearn.model_selection.train_test_splitdocumentation. Controls the shuffling applied to the data before applying the split. Pass an int for reproducible output across multiple function calls. See Glossary.By default
None.- shuffle: bool, default=True
For the
sklearn.model_selection.train_test_splitdocumentation. Whether or not to shuffle the data before splitting. Ifshuffle=Falsethen stratify must be None.By default
True.- stratify: array-like, default=None
For the
sklearn.model_selection.train_test_splitdocumentation. If notNone, data is split in a stratified fashion, using this as the class labels. Read more in the User Guide.By default
None.- **fit_paramsdict
Additional fit parameters.
- Returns
- MapieQuantileRegressor
The model itself.
- predict(X: Union[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]], numpy._typing._nested_sequence._NestedSequence[numpy._typing._array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy._typing._nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]], ensemble: bool = False, alpha: Optional[Union[float, Iterable[float]]] = None, optimize_beta: bool = False, allow_infinite_bounds: bool = False, symmetry: Optional[bool] = True) Union[numpy.ndarray[Any, numpy.dtype[numpy._typing._array_like._ScalarType_co]], Tuple[numpy.ndarray[Any, numpy.dtype[numpy._typing._array_like._ScalarType_co]], numpy.ndarray[Any, numpy.dtype[numpy._typing._array_like._ScalarType_co]]]][source]¶
Predict target on new samples with confidence intervals. Residuals from the training set and predictions from the model clones are central to the computation. Prediction Intervals for a given
alphaare deduced from the quantile regression at the alpha values: alpha/2, 1 - (alpha/2) while adding a constant based uppon their residuals.- Parameters
- X: ArrayLike of shape (n_samples, n_features)
Test data.
- ensemble: bool
Ensemble has not been defined in predict and therefore should will not have any effects in this method.
- alpha: Optional[Union[float, Iterable[float]]]
For
MapieQuantileRegresorthe alpha has to be defined directly in initial arguments of the class.- symmetry: Optional[bool]
Deciding factor to whether to find the quantile value for each residuals separatly or to use the maximum of the two combined.
- Returns
- Union[NDArray, Tuple[NDArray, NDArray]]
NDArray of shape (n_samples,) if
alphaisNone.Tuple[NDArray, NDArray] of shapes (n_samples,) and (n_samples, 2, n_alpha) if
alphais notNone.[:, 0, :]: Lower bound of the prediction interval.
[:, 1, :]: Upper bound of the prediction interval.
- set_fit_request(*, X_calib: Union[bool, None, str] = '$UNCHANGED$', calib_size: Union[bool, None, str] = '$UNCHANGED$', groups: Union[bool, None, str] = '$UNCHANGED$', random_state: Union[bool, None, str] = '$UNCHANGED$', sample_weight: Union[bool, None, str] = '$UNCHANGED$', shuffle: Union[bool, None, str] = '$UNCHANGED$', stratify: Union[bool, None, str] = '$UNCHANGED$', y_calib: Union[bool, None, str] = '$UNCHANGED$') mapie.regression.quantile_regression.MapieQuantileRegressor¶
Request metadata passed to the
fitmethod.Note that this method is only relevant if
enable_metadata_routing=True(seesklearn.set_config()). Please see User Guide on how the routing mechanism works.The options for each parameter are:
True: metadata is requested, and passed tofitif provided. The request is ignored if metadata is not provided.False: metadata is not requested and the meta-estimator will not pass it tofit.None: metadata is not requested, and the meta-estimator will raise an error if the user provides it.str: metadata should be passed to the meta-estimator with this given alias instead of the original name.
The default (
sklearn.utils.metadata_routing.UNCHANGED) retains the existing request. This allows you to change the request for some parameters and not others.New in version 1.3.
Note
This method is only relevant if this estimator is used as a sub-estimator of a meta-estimator, e.g. used inside a
pipeline.Pipeline. Otherwise it has no effect.- Parameters
- X_calibstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
X_calibparameter infit.- calib_sizestr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
calib_sizeparameter infit.- groupsstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
groupsparameter infit.- random_statestr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
random_stateparameter infit.- sample_weightstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
sample_weightparameter infit.- shufflestr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
shuffleparameter infit.- stratifystr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
stratifyparameter infit.- y_calibstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
y_calibparameter infit.
- Returns
- selfobject
The updated object.
- set_predict_request(*, allow_infinite_bounds: Union[bool, None, str] = '$UNCHANGED$', alpha: Union[bool, None, str] = '$UNCHANGED$', ensemble: Union[bool, None, str] = '$UNCHANGED$', optimize_beta: Union[bool, None, str] = '$UNCHANGED$', symmetry: Union[bool, None, str] = '$UNCHANGED$') mapie.regression.quantile_regression.MapieQuantileRegressor¶
Request metadata passed to the
predictmethod.Note that this method is only relevant if
enable_metadata_routing=True(seesklearn.set_config()). Please see User Guide on how the routing mechanism works.The options for each parameter are:
True: metadata is requested, and passed topredictif provided. The request is ignored if metadata is not provided.False: metadata is not requested and the meta-estimator will not pass it topredict.None: metadata is not requested, and the meta-estimator will raise an error if the user provides it.str: metadata should be passed to the meta-estimator with this given alias instead of the original name.
The default (
sklearn.utils.metadata_routing.UNCHANGED) retains the existing request. This allows you to change the request for some parameters and not others.New in version 1.3.
Note
This method is only relevant if this estimator is used as a sub-estimator of a meta-estimator, e.g. used inside a
pipeline.Pipeline. Otherwise it has no effect.- Parameters
- allow_infinite_boundsstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
allow_infinite_boundsparameter inpredict.- alphastr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
alphaparameter inpredict.- ensemblestr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
ensembleparameter inpredict.- optimize_betastr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
optimize_betaparameter inpredict.- symmetrystr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
symmetryparameter inpredict.
- Returns
- selfobject
The updated object.
- set_score_request(*, sample_weight: Union[bool, None, str] = '$UNCHANGED$') mapie.regression.quantile_regression.MapieQuantileRegressor¶
Request metadata passed to the
scoremethod.Note that this method is only relevant if
enable_metadata_routing=True(seesklearn.set_config()). Please see User Guide on how the routing mechanism works.The options for each parameter are:
True: metadata is requested, and passed toscoreif provided. The request is ignored if metadata is not provided.False: metadata is not requested and the meta-estimator will not pass it toscore.None: metadata is not requested, and the meta-estimator will raise an error if the user provides it.str: metadata should be passed to the meta-estimator with this given alias instead of the original name.
The default (
sklearn.utils.metadata_routing.UNCHANGED) retains the existing request. This allows you to change the request for some parameters and not others.New in version 1.3.
Note
This method is only relevant if this estimator is used as a sub-estimator of a meta-estimator, e.g. used inside a
pipeline.Pipeline. Otherwise it has no effect.- Parameters
- sample_weightstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
sample_weightparameter inscore.
- Returns
- selfobject
The updated object.